
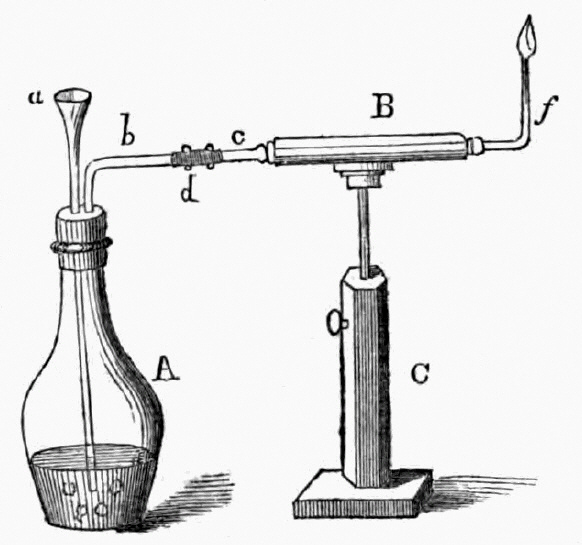
1818 publication by Orfila and Marsh apparatus for the detection of arsenic
a= funnel, b= escape tube, C= stand


| Population of Bangladesh | 125 million |
| Total population in regions where some wells are known to be contaminated | 35-77 million |
| Maximum concentration of arsenic permitted in drinking-water according to WHO recommendations | 10 mg/l |
| Maximum concentration allowed in Bangladesh | 50 mg/l (similar to many countries worldwide) |
| Number of tube-wells sampled by the British Geological Survey (1998) | 2022 |
| Proportion of wells with arsenic concentrations >50 mg/l | 35% |
| Proportion of wells with arsenic concentrations >300 mg/l | 8.4% |
|
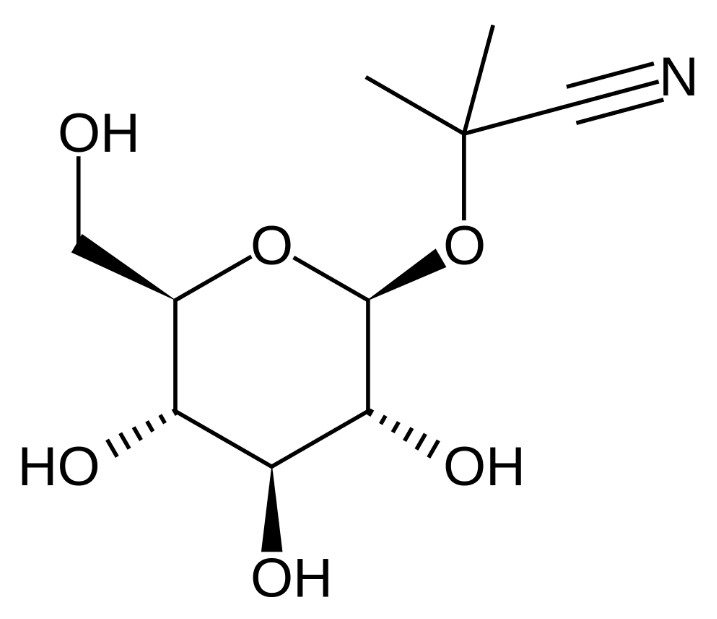
linamarin |
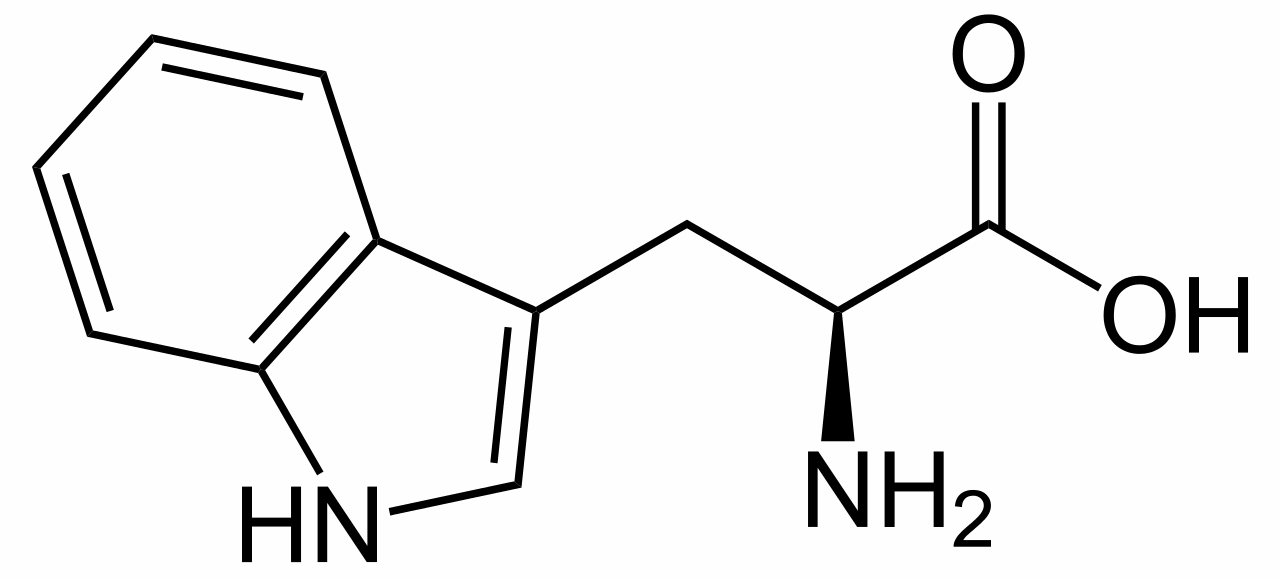
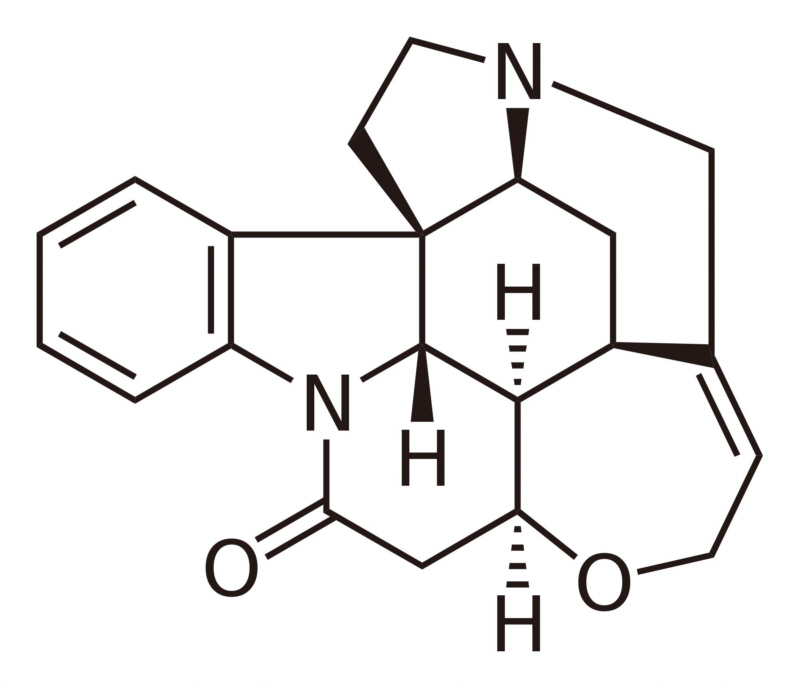 L-tryptophan is the biosynthetic precursor to strychnine |
|
| Toxic Compound | Occurrence | Type | ~RMM | LD50 * |
|---|---|---|---|---|
| Most Toxic | ||||
| maitotoxin | dinoflagellum | polyketide | 3422 | 0.050 |
| ciguatoxin | dinoflagellum | polyketide | 1061 | 0.35 |
| palytoxin | coral species | polyketide | 2679 | 0.45 |
| taipoxin | Australian taipan snake | glycoprotein | 45600 | 2 |
| batrachotoxin | Columbian poison dart frog | steroid alcohol | 539 | 2 |
| tetrodotoxin | pufferfish | saccharide derivative | 319 | 10 |
| Plant poisons | ||||
| ricin | castor bean | glycoprotein (lectin) | 62400 | 0.1 |
| nicotine | tobacco plant | alkaloid | 162 | 300 |
| strychnine | poison nut | alkaloid | 334 | 750 |
| cymarine | tropical creeping shrub | digitalis glycoside | 549 | 25000 |
| tubocurarine chloride | tropical woody vine | alkaloid | 682 | 33200 |
| atropine | deadly nightshade | alkaloid | 289 | 400000 |
| Fungal poisons | ||||
| L-(+)-muscarine | "fly agaric" | alkaloid | 174 | 230 |
| α-amanitin | "death cap" | bicyclic octapeptide | 919 | 300 |
| penitrem A | mould | polycyclic indole derivative | 634 | 1050 |
| aflatoxin b1 | mould | difuran coumarin derivative | 312 | 1700 |
| Inorganic and synthetic poisons | ||||
| 2,3,7,8-TCDD (dioxin) | polychlorinated dibenzo-p-dioxin | 320 | 22 | |
| parathion (E605) | organophoshate | 291 | 3600 | |
| potassium cyanide | 66 | 10000 | ||
| arsenic oxide | 198 | 15100 | ||

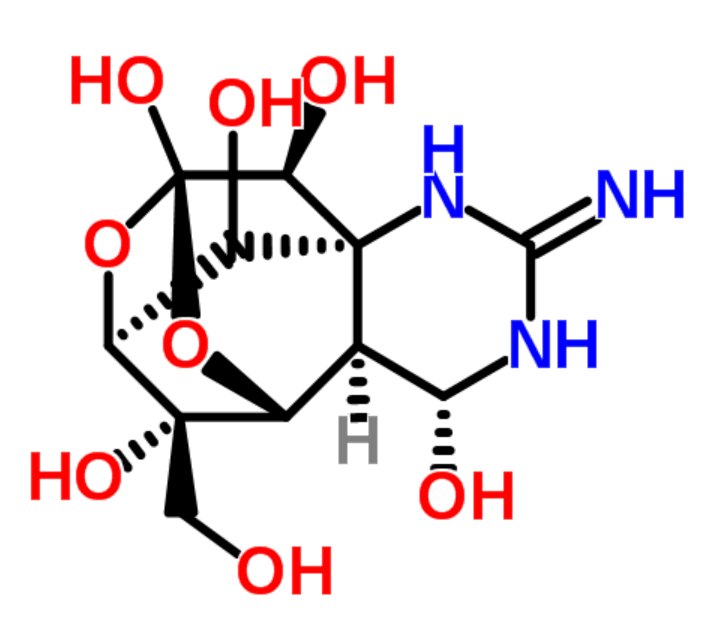
|
tetrodotoxin |
|
epibatidine and batrachotoxin |
|
|
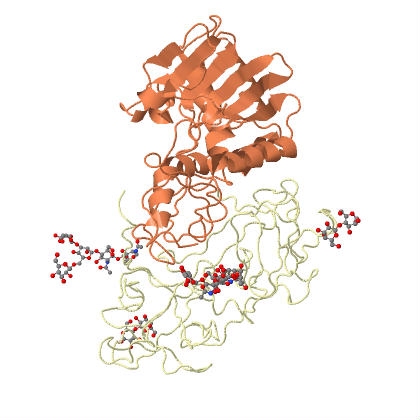
| Ricin is an example of an holotoxin, consisting of heterodimeric glycoproteins, the A and B chains. | |

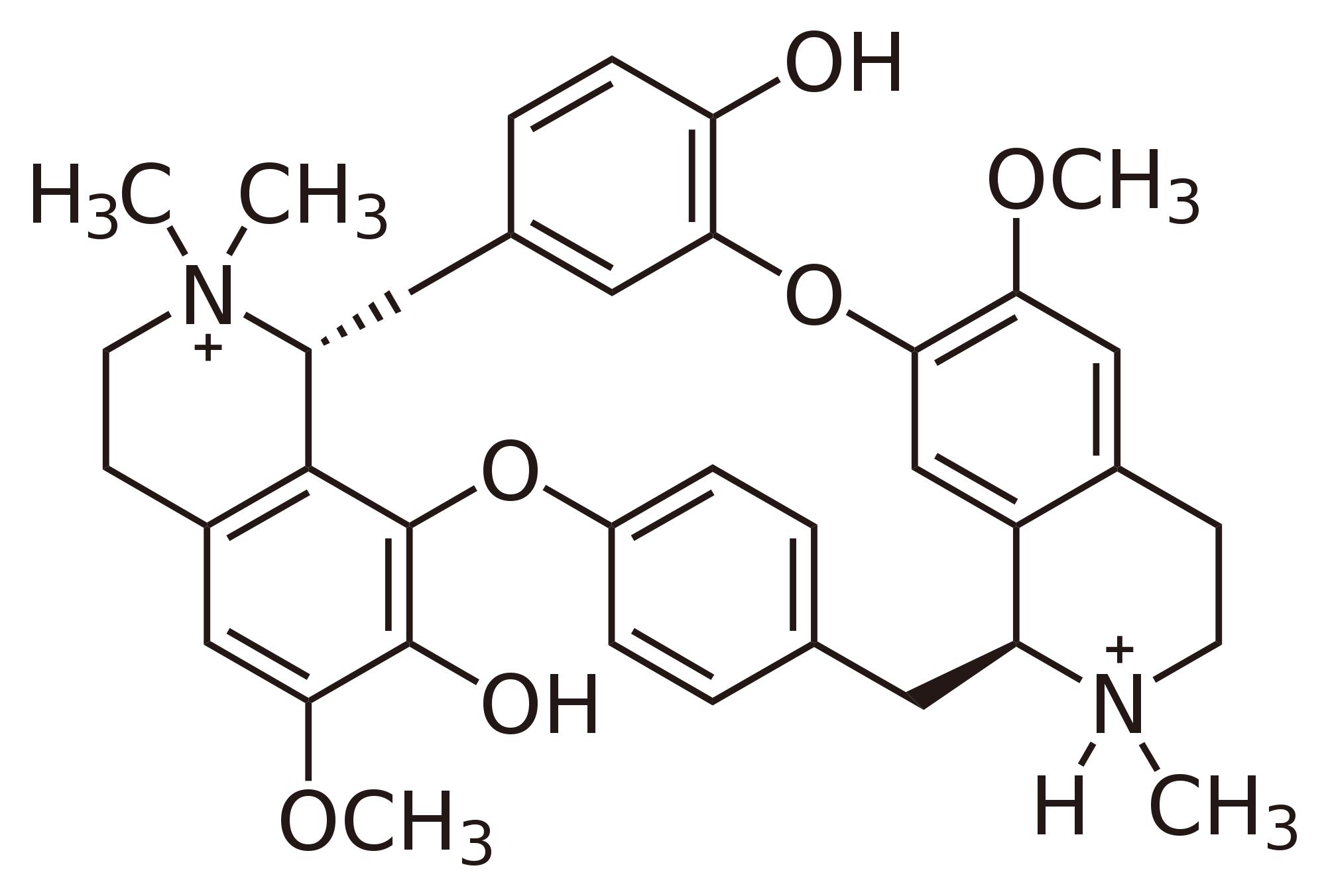
|
tubocurarine |
| Name | R1 | R2 | R3 | R4 | R5 |
|---|---|---|---|---|---|
| α-Amanitin | OH | OH | NH2 | OH | OH |
| β-Amanitin | OH | OH | OH | OH | OH |
| γ-Amanitin | H | OH | NH2 | OH | OH |
| ε-Amanitin | H | OH | OH | OH | OH |
| Amanullin | H | H | NH2 | OH | OH |
| Amanullinic acid | H | H | OH | OH | OH |
| Amaninamide | OH | OH | NH2 | H | OH |
| Amanin | OH | OH | OH | H | OH |
| Proamanullin | H | H | NH2 | OH | H |
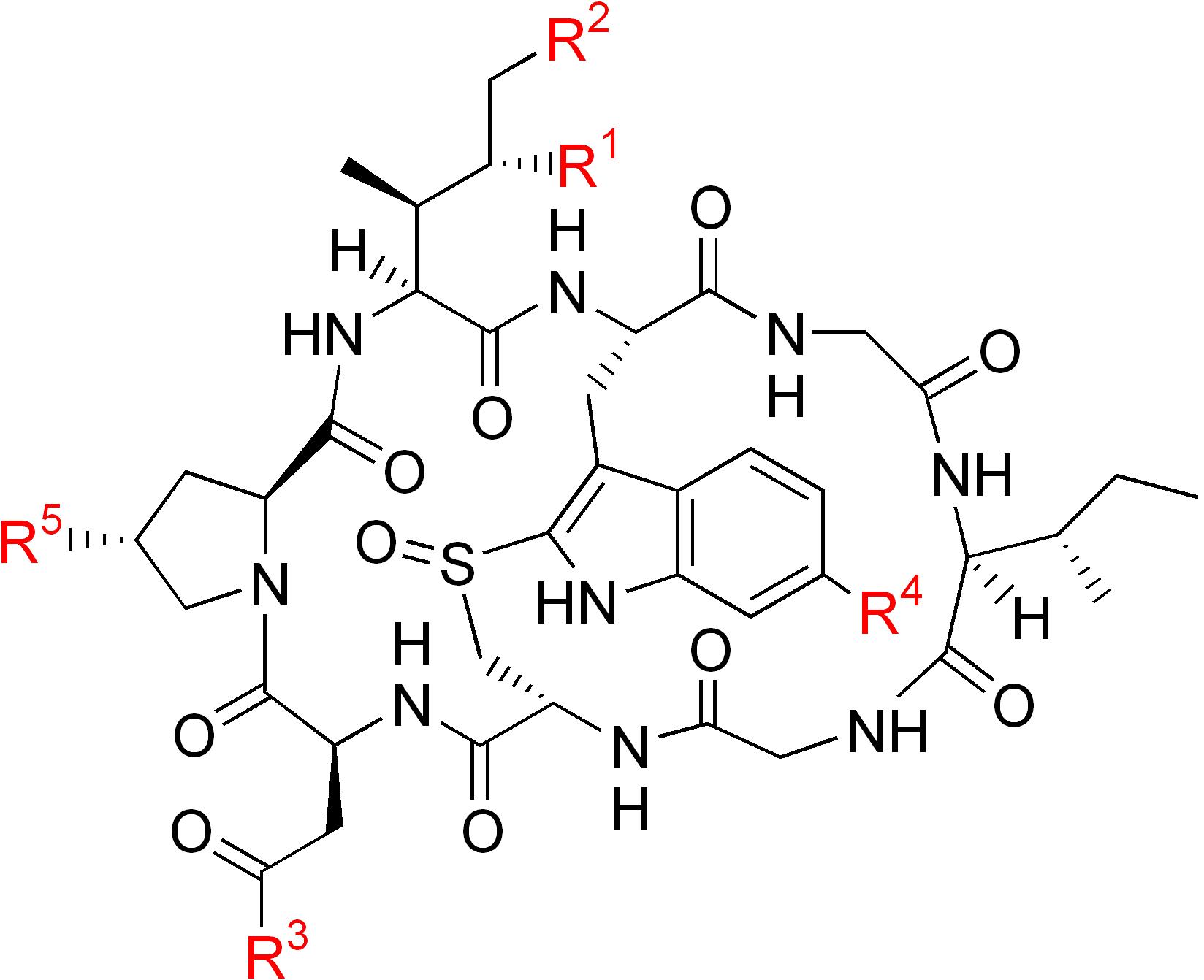
|
α-amanitin |
| Family Name | Plant Name | Common Name |
| Aizoaceae | Trianthema portulacastrum | Horse Purslane |
| Amaryllidaceae | Haemanthus mulitflorus | Blood Lily |
| Apocynaceae | Adeniuzn spp. | Desert Rose |
| Catharanthus roseus | Periwinkle, Rain Goat Rose | |
| Nerium oleander | Oleander | |
| Rauvolfia nitida | Glasswood | |
| Thevetia peruviana | Milk Bush, Yellow Oleander, Lucky Beans | |
| Urechites lutea | Nightsage, Yellow Nightshade | |
| Aristolochiaceae | Aristolochia grandiflora | Poisoned Hog Meat |
| Asclepiadaceae | Asciepias curassavica | Red Head, Red Top, Blood Flower |
| Calotropis procera | French Cotton, Dumb Cotton | |
| Cryptostegia grandiflora | Purple Allamanda, India-rubber Vine | |
| Araceae | Dieffenbachia seguine | Dumb Cane |
| Boraginaceac | Heliotropium indicuni | Scorpion Weed, Wild Clary |
| Caesalpiniaceae | Cassia alata | Ringworm Shrug |
| Campanulaceae | Hippobroma longiflora | Madame Fate, Horse Poison, Star Flower |
| Cannabinaceae | Cannabis sativa | Ganja, Marijuana, Indian Hemp |
| Chenopodiaceae | Chenopodium ambrosioides | Semicontract, Mexican Tea, Bitter Weed |
| Compositae (Asteraceae) | Erechtites hieraciifolia | Fireweed |
| Senecio discolor | Whiteback | |
| Cucurbitaceae | Luffa aegyptiaca | Strainer Vine, Loofah Gourd |
| Euphorbiaceae | Hippomane mancinella | Manchineel Tree |
| Jatropha curcas | Physic Nut Tree | |
| Manihot esculenta | Cassava, Tapioca | |
| Ricinus communis | Castor Oil Plant, Oil Nut Plant | |
| Tragia volubilis | Twining Cowitch | |
| Lauraceae | Cassytha filiformis | Love Bush |
| Liliaceae | Gloriosa rothschildiana | Gloriosa Lily |
| Loganiaceae | Spigeia anthelmia | Pink Weed, Worm Grass |
| Loranthaceae | Phoradendron piperoides | Godbush, Scorn-the-Earth |
| Malvaceae | Sida acuta | Broomweed |
| Meliaceae | Melia azedarach | Neem, China Berry, Persian Lilac |
| Menispermaceae | Cissampelos pareira | Velvet Leaf |
| Mimosaceae | Albizia saman or Samanea saman | Guango, Rain Tree, Cow Tamarind |
| Papaveraceae | Argemone mexicana | Mexican Poppy, Yellow Thistle |
| Papiionaceae | Abrus precatorius | John Crow Bead Vine, Crab's eyes, Red Bead Vine |
| Andira inermis | Cabbage Bark Tree | |
| Crotalaria fulva | Consumption Weed | |
| Mucuna pruriens | Cowitch | |
| Ormosia jamaicensis | Red Nickel | |
| Portulacaceae | Portulaca oleracea | Pussley, Pursiane |
| Punicaceae | Punica granatum | Pomegranate |
| Rubiaceae | Borreria verticillata | Button Weed, Wild Scabious |
| Cinchona pubescens | Quinine Tree | |
| Sapindaceae | Blighia sapida | Ackee |
| Solanaceae | Cestrum diurnum | Wild Jasmine |
| Datura stramonium | Devil's Trumpet, Thorn Apple, Jimson Weed | |
| Nicotiana tabacum | Tobacco | |
| Solanium ciliatum | Cockroach Poison | |
| Verbenaceae | Lantana camara | Whitesage, Wild Sage |
| Alkaloid | Isolation | Structure | Synthesis |
| (yrs to det.) | |||
| Morphine | 1805 | 1925 (120) | 1952 |
| Strychnine | 1818 | 1947 (129) | 1954 |
| Atropine | 1819 | 1883 (64) | 1902 |
| Quinine | 1820 | 1908 (88) | 1944/2003 |
| Caffeine | 1820 | 1882 (62) | 1895 |
| Nicotine | 1828 | 1893 (65) | 1904 |
| Cocaine | 1860 | 1898 (38) | 1898 |

This work is licensed under a Creative Commons
Attribution-ShareAlike 3.0 Unported License.
 Return to Chemistry,
UWI-Mona, Home Page
Created and maintained by Prof. Robert J.
Lancashire,
Return to Chemistry,
UWI-Mona, Home Page
Created and maintained by Prof. Robert J.
Lancashire,